Example 4: Causal discovery in a predictive coding network#
Authors
Lina Walkowiak, Aarhus University, Denmark (202205493@post.au.dk)
Nicolas Legrand, Aarhus University, Denmark (nicolas.legrand@cas.au.dk)
from functools import partial
from typing import Callable, Dict, NamedTuple, Optional, Tuple
import jax.numpy as jnp
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
from jax import Array, jit
from pyhgf.model.network import Network
from pyhgf.typing import Edges
np.random.seed(123)
plt.rcParams["figure.constrained_layout.use"] = True
In this notebook, we are interested in the possibility of dynamic causal discovery in predictive coding networks. Generalised hierarchical Gaussian filters are Bayesian models, and as such, they imply a portion of causality in the framework: a parent node can exert a causal influence on a child node. The strength of this causal influence is controlled by *_couplings_* parameters in the network’s attributes.
This is in a situation where the parent is updated by the prediction errors returned by the children. But causality can also be inferred from variables that remain independent during the learning process - therefore the cause should not be updated based on a change in the effect. This corresponds to a causal discovery principle, and we can define \( \alpha_{1 \rightarrow 2} \in [0, 1]\) the causal strength that describes how much a variable \(X_1\) is influencing another variable \(X_2\). In this tutorial, we explain how to infer this variable dynamically using causal prediction errors.
Simulation#
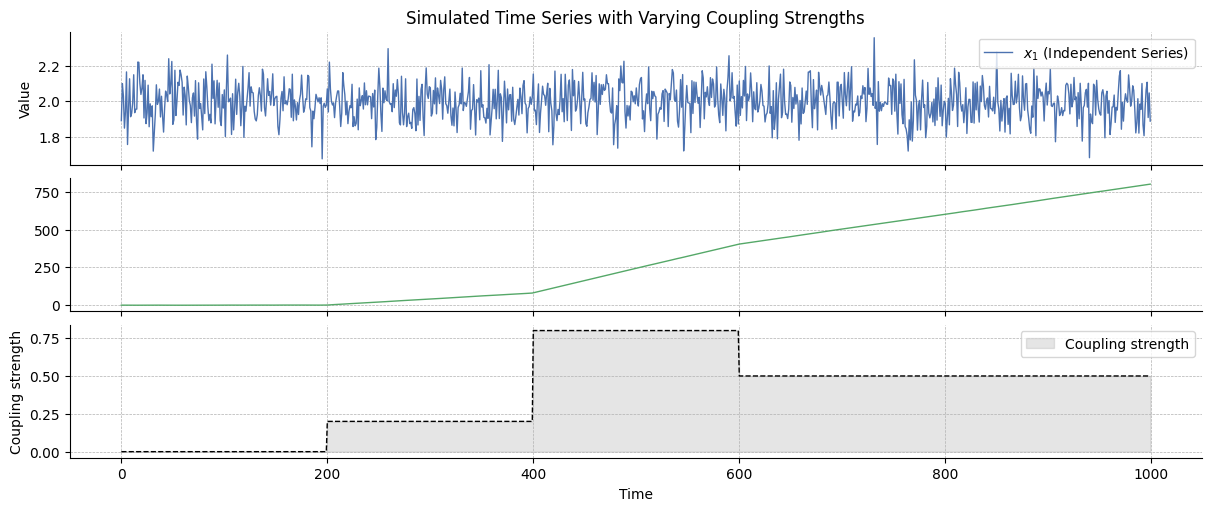
We create two time series, \(x_1\) and \(x_2\), with \(x_1\) influencing \(x_2\) with a an intensity noted \(\alpha_{1 \rightarrow 2} \in [0, 1]\) that is varying over time. Both random variables are Gaussian random walks such as:
We can explicitly inform this model that \(X_1\) influences \(X_2\) from one time step to the next proportionally to a coupling strength, such as:
Given the rule for the sum of normally distributed random variables, we have:
We simulate below two vectors following these principles:
# Parameters
n_samples = 1000
# Generate x_1 and x_2 as a random walk
x1 = np.ones(n_samples) * 2 + np.random.normal(0, 0.1, n_samples)
x2 = np.zeros(n_samples)
# Coupling vector
coupling = np.array([0.0, 0.0, 0.2, 0.2, 0.8, 0.8, 0.5, 0.5, 0.5, 0.5]).repeat(
n_samples / 10
)
# Update x_2 so it is influenced by x_1 according to the coupling vector
for i in range(1, n_samples):
x2[i] = np.random.normal(x2[i - 1] + coupling[i] * x1[i - 1], 0.1)
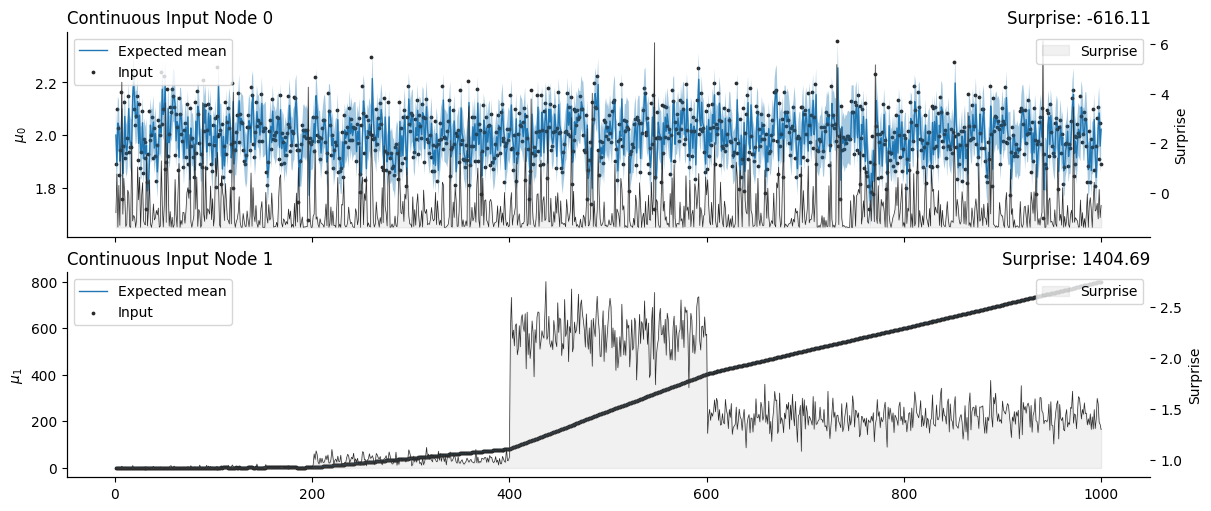
A non-causal network#
We can filter these streams of observation using a simple one-level model without assuming dependencies between the parent nodes or the variables.
# Initialize two independent HGFs for X1 and X2
non_causal_hgf = (
Network()
.add_nodes(precision=100.0)
.add_nodes(precision=1.0)
.add_nodes(value_children=0, mean=2.0)
.add_nodes(value_children=1, tonic_volatility=5.0)
)
non_causal_hgf.plot_network()
# Input the time series
input_data = np.array([x1, x2]).T
non_causal_hgf.input_data(input_data=input_data);
# Plot trajectories for each HGF
non_causal_hgf.plot_nodes(node_idxs=[0, 1])
sns.despine()
Deriving causal prediction errors#
We can also assume that the first input node tries to discover its causal children over time by trying to contribute the the prediction of the other node and learning from their error in doing so. Given a new observation \(u_1\), received by the node \(x_1\), we can define a precision-weighted prediction errors \(\delta_1\) for the non-causal hypothesis, where node \(1\) only use its expectation to predict new incoming values$:
And we can also define a second prediction error \(\delta_{1 \rightarrow 2}\) for the causal hypothesis, this time assuming that \(X_1\) is added to \(X_2\) proportionally to a coupling strength \(\alpha\):
Let \(f(\alpha)\) denote the squared precision-weigthed prediction error when assuming a given \(\alpha\) as:
This function has a first derivative defined as:
Proof. ### Derivation of \(f'(\alpha)\)
The squared precision-weighted prediction error function is defined as: $\( f(\alpha) = (u - \mu_2 - \alpha \mu_1)^2 \cdot \frac{1}{\alpha^2 \sigma_1^2 + \sigma_2^2} \)$
We aim to find the roots of the derivative \(f'(\alpha)\), to identify the critical points where \(f(\alpha)\) is minimised.
Expanding \(f(\alpha)\), we have: $\( f(\alpha) = \frac{(u - \mu_2 - \alpha \mu_1)^2}{\alpha^2 \sigma_1^2 + \sigma_2^2} \)$
By using the quotient rule, the derivative \(f'(\alpha)\) can be written as: $\( f'(\alpha) = \frac{(\alpha^2 \sigma_1^2 + \sigma_2^2) \cdot 2 (u - \mu_2 - \alpha \mu_1) (-\mu_1) - (u - \mu_2 - \alpha \mu_1)^2 (2 \alpha \sigma_1^2)}{(\alpha^2 \sigma_1^2 + \sigma_2^2)^2} \)$
We derive the numerator by simplifying: $\( -2 \mu_1 (u - \mu_2 - \alpha \mu_1)(\alpha^2 \sigma_1^2 + \sigma_2^2) - (u - \mu_2 - \alpha \mu_1)^2 (2 \alpha \sigma_1^2) \)$
Factoring terms: $\( f'(\alpha) = -\frac{2 (u - \mu_2 - \alpha \mu_1) \left[\mu_1 (\alpha^2 \sigma_1^2 + \sigma_2^2) + \alpha \sigma_1^2 (u - \mu_2 - \alpha \mu_1)\right]}{(\alpha^2 \sigma_1^2 + \sigma_2^2)^2} \)$
We want to find the value of \(\alpha\) that minimises \(\delta_{1 \rightarrow 2}\) (\(\delta_2\) being a special case where \(\alpha = 0\)). Two roots can be found at:
Proof. ### Finding the roots of \(f'(\alpha)\)
To identify the critical points of \(f(\alpha)\), we solve for the values of \(\alpha\) where \(f'(\alpha) = 0\). This condition is satisfied when the numerator of the derivative equals zero.
As described above, the numerator of the derivative of the precision-weighted prediction error function \(f(\alpha)\) is given by:
There are two scenarios for \(f'(\alpha) = 0\):
\(u - \mu_2 - \alpha \mu_1 = 0\)
\(\mu_1 (\alpha^2 \sigma_1^2 + \sigma_2^2) + \alpha \sigma_1^2 (u - \mu_2 - \alpha \mu_1) = 0\)
We solve each condition separately to find the roots of \(f'(\alpha)\) as described below.
First root: Solving \(u - \mu_2 - \alpha \mu_1 = 0\)
Rearranging this equation for \(\alpha\), we get:
Thus, for the first root, we have:
Second root: Solving \(\mu_1 (\alpha^2 \sigma_1^2 + \sigma_2^2) + \alpha \sigma_1^2 (u - \mu_2 - \alpha \mu_1) = 0\)
To isolate \(\alpha\), we begin with the simplified equation:
Rearranging terms to isolate \(\alpha\):
Solving for \(\alpha\):
By rewriting the denominator, we get the following for the second root:
It is therefore possible to recover the implied parameter \(\hat{\alpha}\) over the range \([0, 1]\) by returning \(\alpha \in [0.0, \hat{\alpha}_1, \hat{\alpha}_2, 1.0]\) such that \(0.0 <= \alpha <= 1.0\) that minimize the precision-weigthed prediction error \(f(\alpha)\).
Causal coupling model#
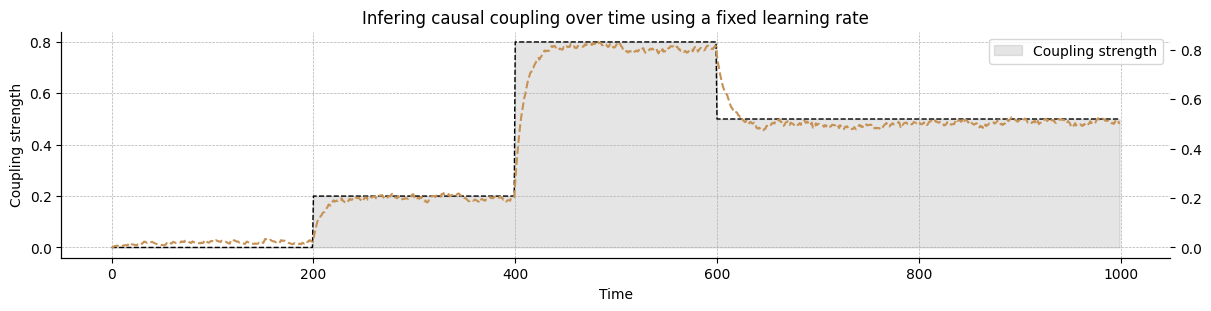
In this section, we create a coupled HGF model to capture the causal influence of the variable \(x_1\) on the variable \(x_2\). This setup now implies learning the strength of a causal connection between \(x_1\) and \(x_2\), which should reflect the actual value used for the simulations above. We therefore use the same model as a starting point and will add an extra step in the update sequence to learn the causal coupling strength over time.
# Initialize a causal HGF
causal_hgf = (
Network()
.add_nodes(precision=100.0)
.add_nodes(precision=1.0)
.add_nodes(value_children=0)
.add_nodes(value_children=1, tonic_volatility=5.0)
)
Add a causal connection between the two nodes#
# Add the coupling strength in the node attributes
causal_hgf.attributes[0]["causal_coupling_children"] = jnp.array([0.0])
# Update the edges variable so it stores the index of the causal child
edges = list(causal_hgf.edges)
adjacency_list = edges[0]
# Create a new adjacency variable for this case
class CausalAdjacencyLists(NamedTuple):
node_type: int
value_parents: Optional[Tuple]
volatility_parents: Optional[Tuple]
value_children: Optional[Tuple]
volatility_children: Optional[Tuple]
coupling_fn: Tuple[Optional[Callable], ...]
causal_children: Optional[Tuple]
causal_adjacency_list = CausalAdjacencyLists(
node_type=adjacency_list.node_type,
value_parents=adjacency_list.value_parents,
volatility_parents=adjacency_list.volatility_parents,
value_children=adjacency_list.value_children,
volatility_children=adjacency_list.volatility_children,
coupling_fn=adjacency_list.coupling_fn,
causal_children=(1,),
)
# Insert the new variable back to the edges
edges[0] = causal_adjacency_list
causal_hgf.edges = tuple(edges)
Create the causal update function#
Now that the variables are in place in the network, we need to create a new update function that will estimate the causal strength between the two variables at each belief propagation.
def prediction_error(u, alpha, mu_1, mu_2, var_1, var_2):
return (u - mu_2 - alpha * mu_1) ** 2 * (1 / (alpha**2 * var_1 + var_2))
def find_alpha(u, mu_1, mu_2, var_1, var_2):
# find root 1
alpha_hat_1 = jnp.where(mu_1 == 0.0, 0.0, -(mu_2 - u) / mu_1)
# find root 2
alpha_hat_2 = jnp.where(
(mu_2 - u) == 0.0, 0.0, (mu_1 * var_2) / ((mu_2 - u) * var_1)
)
# evaluate at 0, 1 and the two possible roots
candidates = jnp.array([0.0, alpha_hat_1, alpha_hat_2, 1.0])
candidates = jnp.where((candidates >= 0.0) & (candidates <= 1.0), candidates, 0.0)
# return prediction errors for all candidates
pe = prediction_error(u, candidates, mu_1, mu_2, var_1, var_2)
return candidates[jnp.argmin(pe)]
@partial(jit, static_argnames=("node_idx", "edges"))
def continuous_node_causal_strength(
attributes: Dict,
edges: Edges,
node_idx: int,
) -> Array:
r"""Update the causal strength between this node and all causal children.
Parameters
----------
attributes :
The attributes of the probabilistic nodes.
node_idx :
Pointer to the value parent node that will be updated.
Returns
-------
attributes :
The attributes of the probabilistic nodes.
"""
# get the expected mean and precision from the causal parent
parent_expected_mean = attributes[node_idx]["expected_mean"]
parent_expected_precision = attributes[node_idx]["expected_precision"]
# set a learning rate for the speed of updating
learning_rate = 0.1
# for all causal children, compute the new causal strength
new_strengths = []
for causal_child_idx, strength in zip(
edges[node_idx].causal_children,
attributes[node_idx]["causal_coupling_children"],
):
# get children's expected mean and precision
child_expected_mean = attributes[causal_child_idx]["expected_mean"]
child_expected_precision = attributes[causal_child_idx]["expected_precision"]
# get a new estimate of alpha
new_alpha = find_alpha(
u=attributes[causal_child_idx]["mean"],
mu_1=parent_expected_mean,
mu_2=child_expected_mean,
var_1=1 / parent_expected_precision,
var_2=1 / child_expected_precision,
)
new_strengths.append(strength + (new_alpha - strength) * learning_rate)
# update the strengths vector
attributes[node_idx]["causal_coupling_children"] = jnp.array(new_strengths)
return attributes
# Add this step at the end of the belief propagation sequence
# Here we simply re-use the previous sequence as template
update_steps = non_causal_hgf.update_sequence.update_steps
update_steps += ((0, continuous_node_causal_strength),)
causal_hgf.update_sequence = non_causal_hgf.update_sequence._replace(update_steps=update_steps)
causal_hgf = causal_hgf.create_belief_propagation_fn()
Fitting data and visualisation#
causal_hgf.input_data(input_data=input_data);
System configuration#
%load_ext watermark
%watermark -n -u -v -iv -w -p pyhgf,jax,jaxlib
Last updated: Fri Oct 03 2025
Python implementation: CPython
Python version : 3.12.3
IPython version : 9.6.0
pyhgf : 0.2.8
jax : 0.4.31
jaxlib: 0.4.31
IPython : 9.6.0
pyhgf : 0.2.8
seaborn : 0.13.2
sys : 3.12.3 (main, Aug 14 2025, 17:47:21) [GCC 13.3.0]
numpy : 2.3.3
matplotlib: 3.10.6
jax : 0.4.31
Watermark: 2.5.0